Frequently Asked Questions
The EMERGE database is a Neo4j-powered graph database designed to store nearly all the data generated by the EMERGE project. This includes text-based data, such as temperature, pH or other measurements. It covers temporal and spatial data. If data cannot be stored natively in the graph database itself (due to file size or format), links referring to the data files can be used in lieu.
For a detailed overview of the database and its capabilities, please see the following manuscript:
Bolduc, B., Hodgkins, S. B., Varner, R. K., Crill, P. M., McCalley, C. K., Chanton, J. P., Tyson, G. W., Riley, W. J., Palace, M., Duhaime, M. B., Hough, M. A., IsoGenie Project Coordinators, IsoGenie Project Team, A2A Project Team, Saleska, S. R., Sullivan, M. B., & Rich, V. I. (2020). The IsoGenie database: an interdisciplinary data management solution for ecosystems biology and environmental research. PeerJ, 8, e9467.
For an updated overview of the EMERGE-DB, please see our 2023 AGU poster:
Hodgkins, S. B., Bolduc, B., Miller, D., Rich, V. I., & EMERGE Biology Integration Institute. (2023). Integrating interdisciplinary data: The EMERGE Database and its broader lessons for data management best practices (IN13B-0565). Presented at the AGU Fall Meeting, San Francisco, CA.
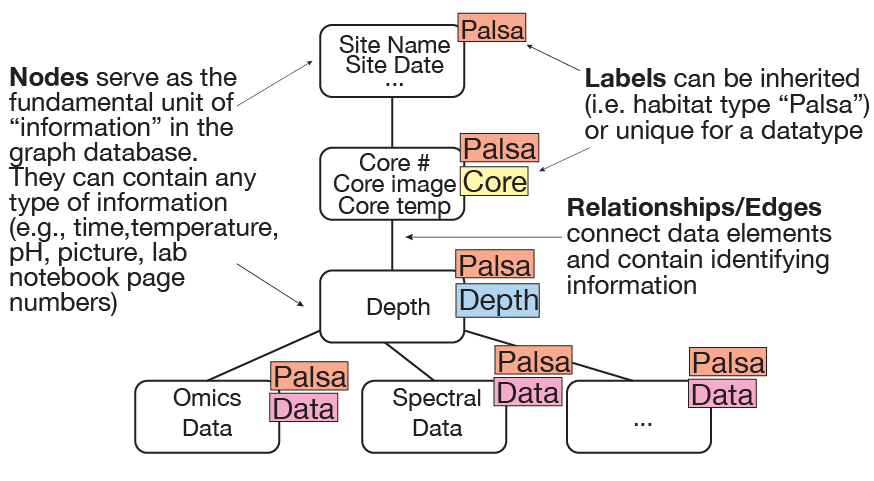
The "database" encompasses several components: (1) the Neo4j graph database, which stores text-based information and serves as the basic storage structure, and (2) the web server, which is built using a number of different tools and provides front-end access to the data contained within and/or linked from the Neo4j database. These two aspects come together to form the EMERGE-DB.
Data are imported into the Neo4j graph database using a set of open-source Python scripts, which are available on our Bitbucket page.
Nearly any type of text-like data can be stored natively in the database. If you check out this article, it turns out the amount of data that can be stored is effectively unlimited. Actual usage is therefore limited by hardware, meaning whatever resources we can purchase. In the EMERGE-DB, text-like data (which includes most coring metadata, biogeochemistry, and timeseries data) is duplicated at various data processing stages, from raw data that can be downloaded at the Data Downloads page, to the data imported into the Neo4j database.
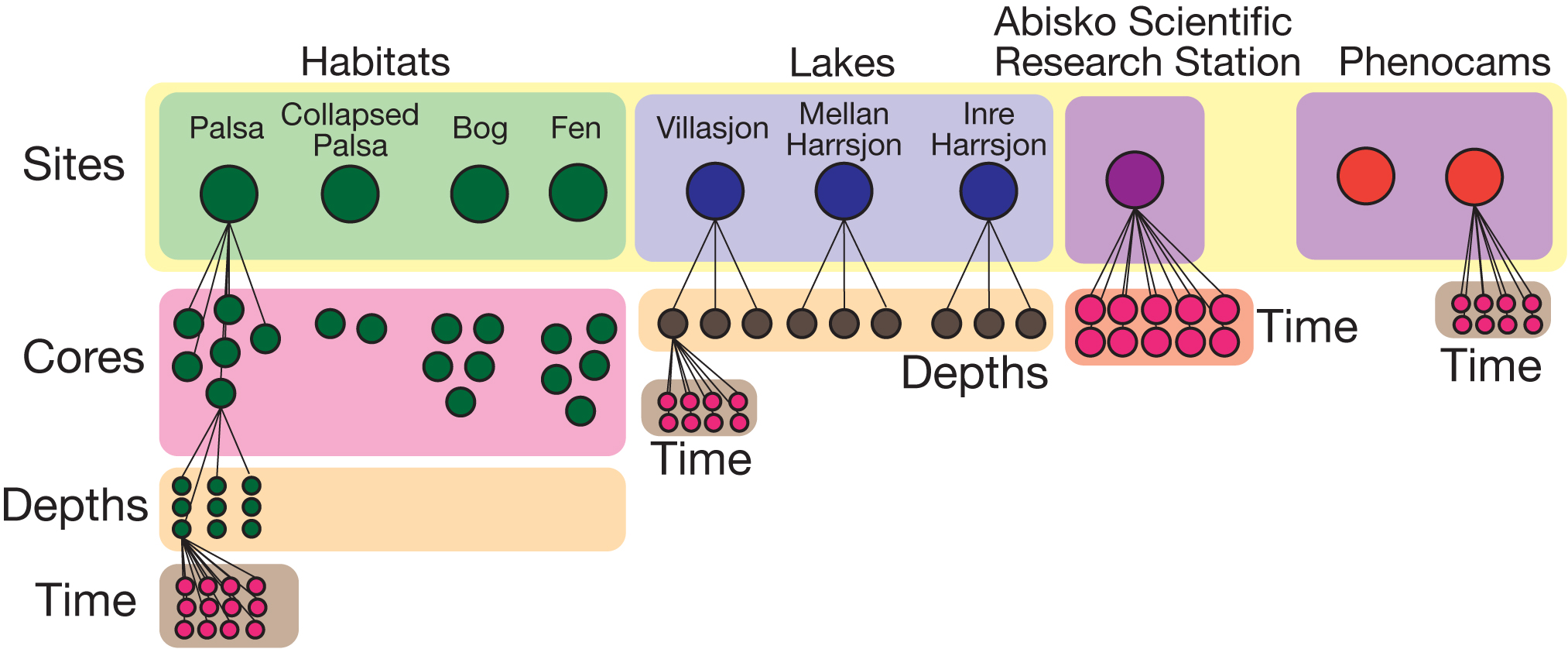
Temporal data can be very high-resolution, e.g. 1 sample/second or 10,000 samples/second. The effective limitation is filtering/aggregating the data so that it is human-comprehensible. For example, let's assume that autochambers take 1 measurement every minute for 5 years straight, that's ~2.628 million measurements. When you query the database, you don't necessarily want to grab all 2+ million nodes, so instead [depending on the observed sample frequencies] the minute measurements are combined into hours or days, reducing the number of nodes to 1,825 (days) or 43,800 (hours). Users can then effectively sort through and filter much larger spans of data more quickly, while still being able to retrieve all the data associated with whatever aggregated level was done.
Images and non-text data, as well as large text files, can have links referring to the actual data locations on the web server or external repositories, allowing users to still access/download the data and see that data when placed in context of other data. This includes most 'omics data (metagenomics, metatranscriptomics, metaproteomics, metabolomics), as well as satellite and drone imagery (both stored as links).
Site synonyms are "unique" terms that different labs use to refer to the same field site. In EMERGE, one example is the use of the words "Sphagnum" and "Bog." One describes the dominant vegetation, the other the habitat classification; but both terms are used by different labs to describe the same "plot of land."
To address this issue, a unified naming scheme was developed to harmonize these divergent names. Whenever data is imported into the database, the site names or their acronyms/abbreviations are passed through a naming dictionary that matches the names against all known variations, and assigns a standardized name for import into the integrated graph database (e.g., for sites, these standardized names correspond to the Site__ property). The dictionary also contains habitat types and other abbreviations which are then associated with the data import. This allows the original data contributors to search for their own data using their own nomenclature, while simultaneously allowing other project members to use the standardized nomenclature to find the same samples.
EMERGE publications include links to the dataset(s) used (either in the EMERGE-DB, or in external repositories linked from the EMERGE-DB) in their "Data Availability" statements. The EMERGE-DB's functionality is intended to survive through the end of the project, and dataset access in perpetuity. As such, references to the website will be maintained, including links from publications to datasets hosted on the legacy IsoGenieDB website (which now redirect to the corresponding pages on this website). In addition, we now use Zenodo to create digital object identifiers (DOIs) for published datasets, ensuring permenant data access long after the project ends; these DOI links are stored in the datasets' Metadata nodes and linked from their pages in the Data Downloads.
The public and private sides of the EMERGE-DB website are integrated into a single framework that shares much of the same content, while still keeping private data isolated from the public side of the website. The public-facing website only provides access to data directed for public release, while the login-protected side of the website provides project members (who must be granted access by a database admin) with access to internally-shared data.
In the Neo4j graph database, only nodes with the "Public" label are accessible from the public Data Downloads and Querying pages, while internal project access for the EMERGE and A2A projects is controlled with the "IsoGenie" and "A2A" node labels (respectively).
While we would ideally like to expand the database to include non-EMERGE project data, we're instead focused on creating a fully comprehensive database for all our members and aren't currently accepting non-EMERGE data.
Querying generally means using a querying language to directly query the underlying Neo4j graph database. As one can imagine, highly sophisticated querying can use not only a query, but also parse and/or retrieve additional information based on the initial data returned. Instead of forcing users to learn a new querying language, we've taken a hybrid approach. Some data is pre-returned to the website for quick filtering. At other times, Python is used in the background to fetch results, translating more common search terms into the querying language used by the Neo4j database. This is especially useful during iterative querying, where prior results affect future queries. For simple queries, i.e. those that have 1 or 2 data types, nearly all data can quickly be filtered via the querying interface.
The major limitation to this method is that there's no easy way of foreseeing what data users will frequently request, or to anticipate exactly what formats are most useful for the end-user. If you find yourself wanting to run the same query repeatedly (simply varying a few search parameters), that can be easily automated.
The EMERGE-DB is spatially-aware in the sense that it contains standardized GPS coordinate information that can be acted upon by other tools/software. This can be seen through the map interface, where GPS information is pulled from the database alongside other site/core-specific information and rendered on a coordinate system.
In a way this means that querying based on GPS information is limited (through the "querying" page) to text-based matches. The Map Interface is another matter. Since GPS information can be retrieved, the limitation to the sophistication of the map queries is limited only by current coding skills and/or available "plugins" that are designed to work with the Google-based mapping software that powers the Map Interface. For example, the drone image on the map is currently overlayed with an installed plugin.
The ultimate goal is a fully embellished, feature-rich Map Interface that combines filtering of site characteristics with summarizing of metadata based on any geospatial selections.